
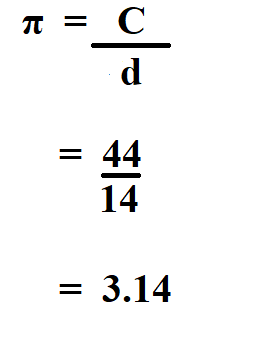
The mean genetic distance among the 861 available pairings of the 42 selected populations was found to be 0.1338. When considering more disaggregated data for 26 European populations, the smallest genetic distance (0.0009) is between the Dutch and the Danes, and the largest (0.0667) is between the Lapps and the Sardinians.
Among this set of 42 world populations, the greatest genetic distance observed is between Mbuti Pygmies and Papua New Guineans, where the Fst distance is 0.4573, while the smallest genetic distance (0.0021) is between the Danish and the English. When studying genetic difference at the world level, the number is reduced to 42 representative populations, aggregating subpopulations characterized by a high level of genetic similarity.įor these 42 populations, Cavalli-Sforza and coauthors report bilateral distances computed from 120 alleles. They focus on aboriginal populations that were at their present location at the end of the 15th century when the great European migrations began. By culling and pooling such samples, they restrict their analysis to 491 populations. Their initial database contains 76,676 gene frequencies (using 120 blood polymorphisms), corresponding to 6,633 samples in different locations. In their study The History and Geography of Human Genes (1994), Cavalli-Sforza, Menozzi and Piazza provide some of the most detailed and comprehensive estimates of genetic distances between human populations, within and across continents. Autosomal genetic distances based on classical markers Larger values are found if highly divergent homogenous groups are compared: the highest such value found was at close to 46%, between Mbuti and Papuans. Within Europe, the most divergent ethnic groups have been found to have values of the order of 7% ( Lapps vs. the Spaniards show values significantly below 1%, indistinguishable from panmixia.

F ST in humans į ST values depend strongly on the choice of populations.Ĭlosely related ethnic groups, such as the Danes vs. The Eastern wolf, a recently recognized highly admixed "wolf-like species" has values of F ST below 10% in comparison with both Eurasian (7.6%) and North American gray wolves (5.7%), with the Red wolf (8.5%), and an even lower value when paired with the Coyote (4.5%). F ST between the Eurasian and North American populations of the gray wolf were reported at 9.9%, those between the Red wolf and Gray wolf populations at between 17% and 18%. Values for mammal populations between subspecies, or closely related species, typical values are of the order of 5% to 20%. Also, strictly speaking F ST is not a distance in the mathematical sense, as it does not satisfy the triangle inequality.įor populations of plants which clearly belong to the same species, values of F ST greater than 15% are considered "great" or "significant" differentiation, while values below 5% are considered "small" or "insignificant" differentiation.

In this case, the probability of identity by descent is very low and F ST can have an arbitrarily low upper bound, which might lead to misinterpretation of the data. The interpretation of F ST can be difficult when the data analyzed are highly polymorphic. If p ¯ is the mutation rate per generation. Two of the most commonly used definitions for F ST at a given locus are based on 1) the variance of allele frequencies between populations, and on 2) the probability of Identity by descent. 5.1 Autosomal genetic distances based on SNPs.5 Autosomal genetic distances based on classical markers.


 0 kommentar(er)
0 kommentar(er)
